ABBA#
ABBA and Brainglobe atlases#
TL; DR
What do I do to configure cuisto properly depending on the atlas and software used ? Check the section below.
Atlases, and especially their coordinates systems, are very important for any downstream processing because both users and softwares need to be aware of which axes correspond to which anatomical directions.
Furthermore, programming language and libraries do not always use the same convention as to how order arrays (3D images numerical representation). Finally, some atlases, such as the 3D version of the mouse spinal cord atlas, are not isotropic (the voxels are not cubic).
Nicolas Chiaruttini, ABBA developper, in a post
Sure, we’ve got all the ingredients for a disaster: the counter intuitive zyx convention for numpy, that has to be mixed with xyz in java, a strange choice of axis for the Allen Brain atlas, and anisotropic atlases. Hopefully we don’t design medical devices...
The following shows the convention used by the Allen Brain Atlas, the Common Coordinates Framework :
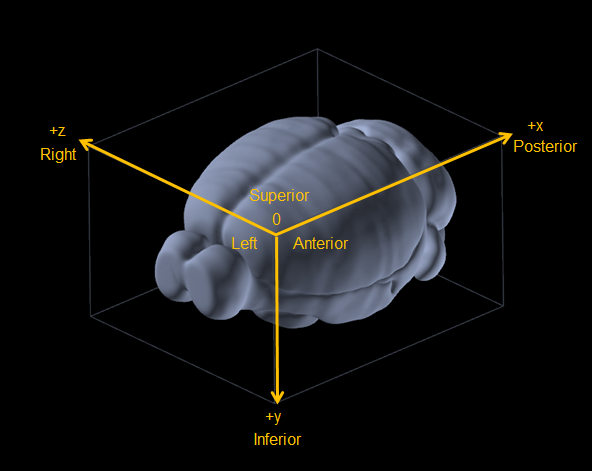
ABBA convention#
Atlases provided by ABBA, namely "Allen Brain Atlas V3p1", follow the CCFv3 convention but slices are assumed to be facing downwards, eg. what you see on the left on the screen is considered to be on the right of the animal. It has the the following consequences :
- Importing registered regions in QuPath will result in regions on the left having the classification "Right: region-name".
- Medio-lateral (left/right) coordinates decreases from left to right.
Point 1. is mitigated in Groovy scripts bundled with cuisto : scripts used to import ABBA registration have an option mirrorLeftRight allowing for Left/Right swap. This is OK as the Allen Brain is symmetrical by construction.
Point 2. is taken into account in cuisto, that detects the hemisphere in which an object is based on the medio-lateral coordinate -- using the atlas "type" parameter in the configuration.
As a side note, ABBA is developped in Java, which uses a x, y, z convention. That means that the first coordinate in ABBA stays the first coordinate in QuPath called Atlas_X and corresponds to the antero-posterior (rostro-caudal) anatomical axis.
Info
Following extensive discussion, yet another atlas was released : "allen_mouse_10um_java". It is the same as the default "Allen Brain Atlas V3p1", but repackaged so it follows the Brainglobe convention. This particular atlas should be treated as a "brainglobe" atlas in cuisto configuration.
Brainglobe convention#
Info
If using regular ABBA from Fiji but using the "allen_mouse_10um_java" atlas, this section applies.
Brainglobe follows the Python/napari/numpy convention for indexing : TCZYX (time, channels, z, y, x). In our case, we can ignore time and channel, keeping the zyx convention. But taking the Allen Brain volume, it considers the antero-posterior direction as \(z\) axis, the dorso-ventral direction as \(y\) axis and the medio-lateral direction as the \(x\) axis.
In other word, for querying the region of a given location using the structure_from_coords() method which can be used like this :
(i, j, k) is a triplet of coordinates :
iis axis 0, eg. rostro-caudal,jis axis 1, eg. dorso-ventral,kis axis 2, eg. medio-lateral.
This looks like CCFv3 and thus ABBA convention, but it deviates in that the axes names are :
zis axis 0, eg. rostro-caudal,yis axis 1, eg. dorso-ventral,xis axis 2, eg. medio-lateral.
This causes a mismatch when using a Brainglobe atlas in ABBA through abba_python. The consequences are :
- First coordinate "Altas_X" is the medio-lateral axis.
- Third coordinate "Atlas_Z" is the rostro-caudal axis.
- The medio-lateral coordinate, "Atlas_X", increases from left to right.
Those points are all taken into account in cuisto using the atlas "type" parameter in the configuration.
Warning
No matter the atlas nor the software (ABBA-Fiji or ABBA-Python), ABBA will always consider slices to be facing downward, thus inverting the Left/Right hemispheres for annotations only, but not necessarily the objects' atlas coordinates.
cuisto configuration#
The main cuisto configuration file has an [atlas] section, in which there is a type parameter.
[atlas] # information related to the atlas used
name = "allen_mouse_10um" # brainglobe-atlasapi atlas name
type = "abba" # abba or brainglobe : registration done with regular ABBA (except allen_mouse_10um_java) or abba_python)
midline = 5700 # midline coordinates in microns (left/right limit, Z for abba, X for brainglobe)
outline_structures = ["root", "CB", "MY", "P"] # structures to show an outline of in heatmaps
Configure like so :
- If using the "Allen Brain Atlas V3p1" atlas, set
type = "abba". - If using any Brainglobe atlases from abba_python, or the "allen_mouse_10um_java" atlas, set
type="brainglobe".
In any event, when importing atlas regions into QuPath with the ABBA extension from the scripts located in scripts/qupath-utils/atlas, set mirrorLeftRight to true. This ensures the regions (annotations) are correctly classified with the correct hemisphere (eg. the left hemisphere is the left part of what you see on the screen).
The type parameter is read to handle axes names ("Atlas_X" and "Atlas_Z"), determine how to extract detections hemisphere and how to plot spatial distributions.
Here is a summary table :
| \ | IJ + Allen Brain Atlas V3p1 | IJ + allen_mouse_10um_java | python + allen_mouse_10um | python + allen_mouse_10um_java |
|---|---|---|---|---|
| First coordinates X | AP | ML | ML | ML |
| Third coordinates Z | ML | AP | AP | AP |
| Side when atlas medio-lateral < 5700 | right | left | left | left |
| Annotations side on left | right | right | right | right |
mirrorLeftRight for Annotations |
true | true | true | true |
cuisto configuration atlas type |
"abba" | "brainglobe" | "brainglobe" | "brainglobe" |